ISCI-1
The ISCF brought together an expert working group to lead its international initiative to draw up globally agreed criteria for characterising stem cell lines. The project was planned to include characterising stem cell lines not currently listed in the US National Institutes of Health (NIH) stem cell registry, and focused on embryonic stem (ES) cell lines.
Our working group included experts in stem cell research and banking from Australia, Canada, Israel, Sweden, the USA and the UK, and liaised closely with the Stem Cell Characterisation Unit at the NIH.
Determining the key criteria for characterisation
The characterisation project involved a comparative study of the different human ES cell isolates that had been collected worldwide. It was expected that the results would enable the working group to reach a consensus about the key criteria that should be used to identify human ES cells, and to establish the degree of heterogeneity that may arise because of different genotypes, different isolation and culture protocols, or because of long term adaptation to culture.
Read the Nature Biotechnology 2005 Commentary on the ISCI project
(LINK to Nature Biotechnology 2005 Commetary)
Read the project overview for details of how the project was organised worldwide and how the stem cell lines were characterised. The conclusions are now published in Nature Biotechnology (atricle available to read) and results are available in the ISCI Registry which also includes detailed protocols used in the different parts of the study (LINK).
Project Overview
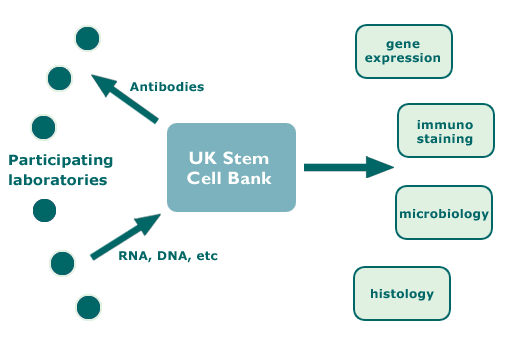
The characterisation project was organised on a ‘hub-and-spoke’ principle. The hub, the UK Stem Cell Bank, collected and prepared antibodies in research-grade facilities for distribution to participating laboratories throughout the world. These laboratories will use the antibodies in specified assays of their cell lines under defined conditions. With agreement of the Institutions owning the different hybridomas, this hybridoma collection at the UK Stem Cell Bank provides an archive stock of key reagents for future reference.
The laboratories provided the hub with cell samples (eg, DNA, RNA and protein, as well as xenograft tumour specimens and fixed cells for immunostaining) according to a specified protocol. The hub then sent the samples to central reference laboratories which undertook to assess the expression of selected genes, provide a DNA fingerprint and to assess microbiological status. Several specialist reference laboratories were engaged to carry out specific assays.
For each stem cell line the plan was to:
1. Establish the expression patterns of
- selected surface antigens
- genes marking undifferentiated stem cells and specific lineages of differentiation.
2. Compare the changes in marker expression in response to a simple differentiation protocol.
3. Establish the degree of correlation between expression of different potential markers of undifferentiated ES cells.
4. Assess its ‘epigenetic’ status with respect to genes subject to imprinting.
5. Provide a DNA fingerprint of each line.
6. Assess its microbiological status, particularly with respect to endogenous retroviruses.
7. Assess the histology of its xenograft tumours.
8. Collate information regarding origins, karyotype, culture conditions, etc.
Basic experimental design
- Cells cultured under ‘local’ conditions
- Cells cultured under ‘standard’ conditions: DMEM/F12 plus 20% Serum Replacement + bFGF
- Assay at 2 time points at least one month apart
- Antigen and Gene expression carried out at same time point
Key project stages
- UK Stem Cell Bank acquires and prepares antibodies for distribution to participating laboratories
- Recruitment of participating laboratories.
- Experimental work.
- Analysis of results.
- ISCF workshop to discuss results and reach consensus on conclusions, followed by publication on ISCF online registry.
Diagram of Hub and Spoke Activities
Participating Laboratories
The participating laboratories fell into two groups: those who had derived their own lines, referred to as “primary laboratories”, and those who had obtained their lines from elsewhere, referred to as “secondary” laboratories. You can browse the stem cell lines by the laboratories who worked on them.
The ISCF working group made the final decision about which laboratories and cell lines were included in the project, to ensure that the scale of the study was kept within manageable bounds and to balance the number of cell lines from different sources.
Primary Laboratories
These laboratories carried out all designated experiments with live cells.
They were asked to:
- Certify that the cells were derived under agreed ethical criteria.
- Agree that data obtained in the initiative will be made public
- Absolve the forum and participating laboratories from legal liability for any outcome of the study
Secondary Laboratories
By involving secondary laboratories we planned to gain some insights into how robust and consistent results were when the same cell lines were cultured by different laboratories.
Pro forma agreement for participants
All data generated by the project is now publicly available on this website. A pro forma agreement was developed for participating laboratories to sign covering: owner permission; ethical standards of cell line derivation; legal liability; and acknowledgement that data generated by the project would be made publicly available.
Central Facilities
UK Stem Cell Bank
The UK Stem Cell Bank and the UK National Institute for Biological Standards (NISB) provided the following services:
Acting as a central receiving ‘hub’
Received materials from participating laboratories and distributed to analytical labs. All material was routed via NIBSC for encoding, which importantly enabled interpretation to be performed blind, with the identity of individual cell lines remaining confidential.
Microbiological analyses
Undertook assays for mycoplasma, known pathogens and endogenous retroviruses.
| Antibody (Antigen) | Marks: | Source/Owner | Reference (details here) |
| TRA-1-60 | hES Cells | Wistar | Andrews et al. 1984b |
| TRA-1-81 | hES Cells | Wistar | Andrews et al. 1984b |
| GCTM2 | hES Cells | ESI/ Pera | Pera et al. 1988 |
| MC631 (SSEA3) | hES Cells | Wistar | Shevinsky et al. 1982 |
| MC813-70 (SSEA4) | hES Cells | Wistar | Kannagi et al. 1983 |
| TRA-2-49 (L-ALP) | hES Cells | Wistar |
Andrews et al. 1984a |
| TRA-2-54 (L-ALP) | hES Cells | Wistar | Andrews et al. 1984b |
| TG30 (CD9) | hES Cells | Pera | |
| Thy1* | hES Cells | Fabre/ICH London | McKenzie and Fabre 1981 |
| MC480 (SSEA1) | Diff Cells | ATCC | Solter and Knowles 1978 |
| VINIS56 (GD3) | Diff Cells | Wistar | Andrews and Oliver 1990 |
| VIN2PB22 (GD2) | Diff Cells | Wistar | Andrews et al. 1990 |
| B159 (NCAM) | Diff Cells | Wistar | |
| W6/32 (HLA) | hES Cells | ATCC | Barnstable et al. 1978 |
* For the ISCI1 project, anti-Thy1 from this hybridoma was provided by Chemicon
Monoclonal antibodies
- Maintainance of reference stocks of hybridomas (see table below)
- Preparation of a reference panel of key antibodies to surface antigens
Reference cell line
Maintainance of a reference stock of 2102Ep human EC cells as a standard for key surface antigens of undifferentiated human ES/EC cells
Other Central Activities
Gene expression patterns
A central analytical laboratory carried out gene expression testing and DNA fingerprinting.
- Markers of undifferentiated ES cells, eg: Oct4, Nanog, Sox2, TDGF (CRIPTO), FoxD3
- Markers of differentiated lineages
A selected panel of 93 genes was analysed by the ABI Microfluidics Real Time RT.PCR system.
In situ immunostaining- M. Pera, Monash University
The spatial distribution of cell types in stock cultures was assessed.
Histopathology - I Damjanov, University of Kansas
Xenograft tumour histology of submitted slides was assessed.
Epigenetic studies - R. Pedersen, P Rugg-Gunn, University of Cambridge
The expression patterns of several genes that should be expressed monoallelically in ES cells was tested.
Statistical analyses- G. Churchill, B. King, J. Szatkiewicz, W. Zhang, The Jackson Laboratory
The data was analysed to address the questions of whether subsets of hES cells can be identified and which markers provide the best correlates of an undifferentiated hES phenotype.
Results of ISCI1
The results obtained from ISCI1 now form part of the ISCI Stem Cell Registry.
